
Download – Source Code – Manual – Preprint – Paper
OrganL is a stand alone Monte Carlo simulation toolkit for the study of complex membranes and subcellular systems. It is under ongoing development. It is able to model
- Complex Lipid Mixtures and Simple Protein-Mimicks
- Arbitrary 3D Geometries
- Open Meshes and Edge Energies
- Constaints and Complicated Boundary Conditions
- Classical Helfrich, Surface Tension and ADE functionals
- etc.
Example Outputs and Videos
Download – Source Code – Manual – Paper
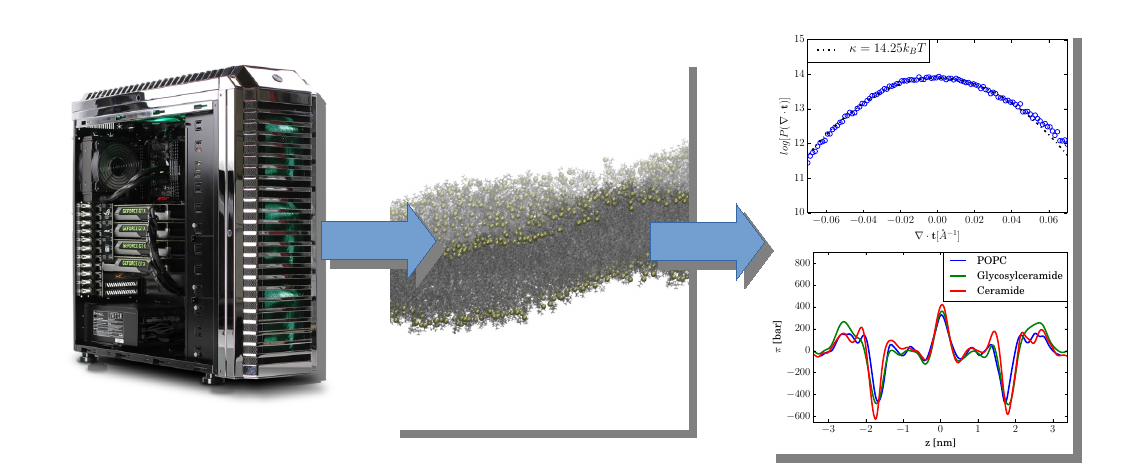
Lipidator Toolkit is a set of analysis tools for MD trajectories. The trajectories are expected to come in the GROMACS xtc file format. This tool can extract local bending rigidity and tilt moduli for arbitrary membrane simulations, using the ReSIS method of which it is the reference implementation. It also has the following additional features:
- Compute bending, tilt and twist moduli from director fluctuations in Fourier space using Frank brown’s method
- Library of director definitions for common lipids compatible with CHARMM, MARTINI and Slipids
- Height fluctuation analysis
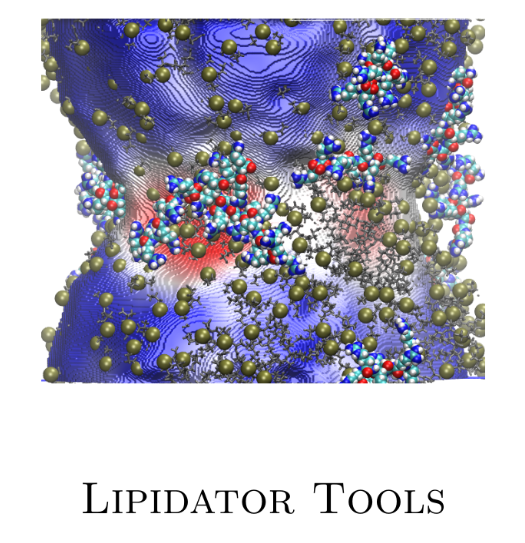
- Generating Gaussian cube files of membrane geometry with curvature maps, as shown on the left image
- An alternative implementation of the tilt modulus from flat surfaces according to G. Khelashvili
For example inputs contact the PI. Source Code Paper
GROMACS Virial version is a fork of the GROMACS simulation toolkit which implements access to the local stress tensor using a Goetz-Lipowski (GLD) force decomposition, based on a PME-Harasima contour implementation by Marcello Sega. The version in this link is compatible with the Sega version and should handle the same way.
The advantage of the GLD is its numerical stability, which is of paramount importance when using e.g. the CHARMM forcefield.
This method can be combined with the lipidator toolkit to get detailed-local continuum properties from membranes and other thin layer systems. It can generate surface tension profiles.